Welcome to Fiber Analysis Website
DNA fiber Analysis allows one to gain critical knowledge of DNA metabolism at the single molecule level. From its “humble” beginnings of being a way to study DNA replication in E.coli, it has blossomed into being a way for researchers to gain critical insights into DNA metabolism (replication, DNA repair), intra-S checkpoint activation, G1/S and S/G2 transitions, chromatin remodeling and modification, and etc.
DNA fiber analysis (be it with DNA or Chromatin fibers) can take a long time to master the methods used to generate these single molecules, to score and measure the various molecules, and to analyze the results. To help facilitate an investigator in learning these techniques or to rapidly obtain results, the DNA fiber core facility will work alongside an investigator so either or both can occur.
We have over 10 years experience helping researchers’ master DNA fiber analysis and these efforts have produced countless manuscripts and have helped numerous grants get funded.
Our Mission
To provide researchers DNA fiber analysis toolset that enables them to gain critical and novel molecular insights into biological processes which will ultimately allow methods to be developed to prevent disease, improve disease detection, and to improve the chances of a positive response towards a treatment regimen.
About Dr. Paul Chastain
Dr. Paul Chastain earned his B.S. in Biochemistry from Indiana University and his Ph.D. in Biochemistry and Biophysics from Texas A&M University. He spent five years as a Lineberger Comprehensive Cancer Center Fellow and three years as an Environmental Pathology Fellow at the University of North Carolina before joining the Department of Pathology and Laboratory Medicine at UNC as an Independent Investigator. His research focused upon determining the mechanism by which oxidative, UV- and chemical carcinogen-induced damage or DNA instability influence DNA replication, and how that disruption may increase the likelihood of disease onset. His researched was funded by National Institutes of Health (NIH), National Institute of Environmental Health Sciences (NIEHS), National Cancer Institute (NCI) and pilot project grants from the Center for Environmental Health and Susceptibility in the UNC-Chapel Hill. Dr. Chastain had the honor of being an Assistant Professor of Biochemistry at William Carey University College of Osteopathic Medicine (WCUCOM) from 2013 to 2016. He primarily co-taught with Professor Robert Bateman Medical Biochemistry to first year medical students and developed mannequin based clinical simulations. In 2016, he joined the medical education department at University of Illinois College of Medicine at Rockford. He is helping to develop the new medical curriculum, teaching medical biochemistry and continues to develop clinical simulations. Research wise, he is researching how the internal and external environment leads to disease formation and progression. He published twenty-nine scientific publications, 1 book chapter, two software packages, and one patent.
His research interests include:
- determining the mechanisms (e.g., intra-S checkpoint, repair pathways) by which the cell protects itself from environmental and endogenous agents (e.g., UV radiation, chemical carcinogens, oxidative stress, environmental pollutants) and how disruption of these protective mechanism increases the chance of cancer and disease formation;
- developing, writing, and implementing computational and mathematical tools (based on Matlab, ImageJ, SQL, C++, and Java) to store, retrieve and analyze biological data and gain critical knowledge from patient specimens.
More information about Dr. Chastain see on his personal website.
Fiber Analysis
History of Fiber Analysis
DNA fiber autoradiography. The generation of DNA fibers to study replication was originally devised by Cairns in 1963 to study DNA replication in E.coli. In this original method thymidine was used to label the DNA and Millipore filters were the medium which supported the fibers. In 1968 Huberman and Riggs adapted the method to allow for the visualization of multiple replicons of mammalian chromosome (Oxford Reference). The knowledge gained from these three pioneers has been instrumental in our knowledge about all aspects of DNA metabolism.
DNA fiber spreading. In 1993 Parra and Windle developed a method which uses a glass surface to capture DNA fibers directly from cells that were lysed. From their paper “the stretching of duplex DNA was accomplished by lysing cells with detergent at one end of a glass slide and allowing the DNA in solution to stream hydrodynamically down the slide”. They also used fluorescent hybridization to map specific locations within the fibers.
Molecular Combing. In 1994 Bensimon et al developed a method to align and straighten purified DNA using a moving interface. To detect the DNA, they used a fluorescent DNA intercalator. In 1997, the method was further developed by Bensimon in which silanized cover slips are dipped into a solution containing DNA and then mechanically pulled out of the solution.
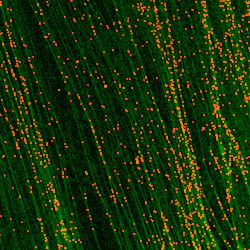
Pulse Labeling with Fluorescent Nucleotides. In 1998 Jackson and Pombo used DNA fiber spreading along with consecutive pulse labeling of iododeoxyuridine (IdU) and chlorodeoxyuridine (CldU), two thymidine analogs, to study nascent DNA synthesis during the time of the pulses. In order to visualize DNA fibers which harbored the IdU and CldU pulses (IdU and CldU tracks), fluorescent antibodies against those analogs were used.
Replication Stress. In 2004, Merrick, Jackson, and Diffley used DNA fiber spreading and pulse labeling before and after various replication stress agents to determine how those agents influenced various replication intermediates (origin initiation fork elongation).
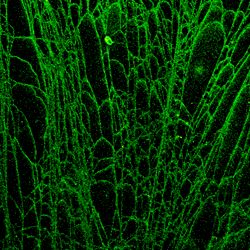
Chromatin Fibers. In 2010, Cohen and Chastain developed a method which allowed chromatin fibers to be prepared which allowed one to visualize not only nascent DNA synthesis by using thymidine analogs but also to visualize the relative location of various DNA metabolism proteins, chromatin remodelers, and chromatin modifications.
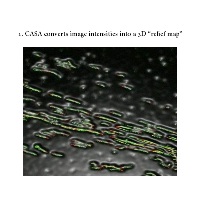
Automated DNA Fiber Analysis. In 2011, Wang and Chastain developed software called computer aided scoring and analysis (CASA) which automated the process of tracking and measuring DNA fibers, nascent DNA synthesis, and the distribution of various areas of interest (DNA damage, location of DNA metabolism proteins, etc.) within those regions.
Some Laboratories who are currently or have used CASA for their research
- Carolyn Price, University of Cincinnati, USA
- Joe Pomerening, Indiana University, USA
- Geoffrey Kapler, Texas A&M University, USA
- Samuel H. Wilson, National Institute of Environmental Health Sciences (NIEHS), USA
- Jason Stewart, University of South Carolina, USA
- Andrew Burgess, ANZAC Research Institute, Australia
- Basuthkar J. Rao, TaTa Institute of Fundamental Research, India
- Keshav K. Singh, University of Alabama at Birmingham (UAB) Med School, USA
- Robert Sobol, USA Mitchell Cancer Institute, University of South Alabama, USA
- Stephen DeVito, Georgetown University, USA
Services
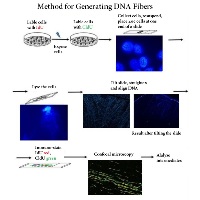
DNA fiber analysis provides a wide platform of services that can be tailored to suit the needs and wishes of a researcher. DNA fiber analysis can be broken down into four parts and thus our services are broken down around those steps:
The first step in DNA fiber analysis is the preparation of DNA fibers. We have streamlined our approach such that one can learn DNA fiber spreading and become an “expert” in less than 1 hour (supplies only cost about $50). If we come into your lab in the morning, you will be an expert by the afternoon.
The second step is visualization of DNA fibers. We have methods for visualizing DNA fibers that are relatively straightforward (you can find the protocol on this website) and easy to do/learn (if you do a western blot to visualize whether your protein of interest is present or not, you can visualize DNA fibers). Visualization of DNA fibers manually takes about 5 – 6 hours to stain about 10 slides at a time (due to the use of multiple antibody layers to visualize replication intermediates of interest) and about 4 hours to stain 30 slides with the help of a slide stainer (we programmed an automatic slide stainer with parameters that were needed to stain the fibers and it worked beautifully). The material cost per slide to visualize DNA fibers manually is a few dollars and a little more to do it using an automatic stainer.
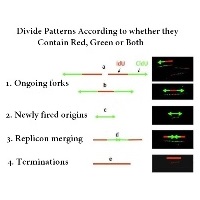
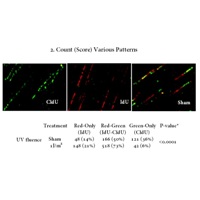
The third step is the analysis of the fibers to obtain the knowledge you are interested in gaining. We have a program that does the analysis for you so this step is the easiest! See software section for more information.
The last step is determining what the data means. This step is the fun step and with our approach you can be at this step 3-4 working days after your experiment!
Since we have been doing this for such a long time, we are well versed in how to prepare DNA fibers, visualize them, and analyze them – therefore, we are comfortable teaching all aspects of this process or being used as a consultant just in case one runs into hiccups along the way. We have been doing DNA fiber analysis and single molecule analysis for over 15 years now (we started doing this type of work in 1997) and we have encountered pretty much all the problems one could face – so we can typically help people resolve their technical issues relatively fast (in one case we helped someone out in less than 1 minute!). We have traveled throughout the US and Europe consulting and teaching DNA fiber analysis so we know that our approach and advice are golden.
Contact
Mailing Address
Dr. Paul David Chastain
Assistant Research Professor
University of Illinois College of Medicine at Rockford
Department of Health Sciences Education
1601 Parkview Avenue, Rockford, Illinois 61107
Telephone:815-395-5622
Email: pdchast7@uic.edu